100 assays (manual) / 1000 assays (microplate) / 1300 assays (auto-analyser)
Prices exclude VAT
Available for shipping
| Content: | 100 assays (manual) / 1000 assays (microplate) / 1300 assays (auto-analyser) |
| Shipping Temperature: | Ambient |
| Storage Temperature: | Short term stability: 2-8oC, Long term stability: See individual component labels |
| Stability: | > 2 years under recommended storage conditions |
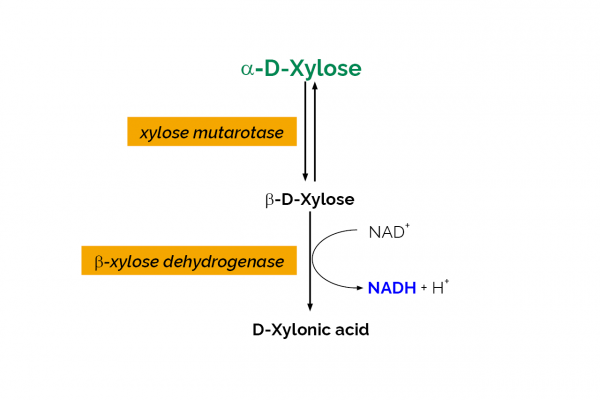
| Analyte: | D-Xylose |
| Assay Format: | Spectrophotometer, Microplate, Auto-analyser |
| Detection Method: | Absorbance |
| Wavelength (nm): | 340 |
| Signal Response: | Increase |
| Linear Range: | 2 to 100 μg of D-xylose per assay |
| Limit of Detection: | 0.7 mg/L |
| Reaction Time (min): | ~ 6 min |
| Application examples: | Analysis of D-xylose in fermentation broths and hydrolysates of plant material and polysaccharides. |
| Method recognition: | Novel method |
The D-Xylose test kit is a novel method for the specific, convenient and rapid measurement and analysis of D-xylose in plant extracts, culture media/supernatants and other materials.
Note for Content: The number of manual tests per kit can be doubled if all volumes are halved. This can be readily accommodated using the MegaQuantTM Wave Spectrophotometer (D-MQWAVE).
View our full range of monosaccharide assay kits.

- Very cost effective
- All reagents stable for > 2 years after preparation
- Only enzymatic kit available
- Rapid reaction (~ 6 min)
- Mega-Calc™ software tool is available from our website for hassle-free raw data processing
- Standard included
- Suitable for manual, microplate and auto-analyser formats
Measurement of available carbohydrates in cereal and cereal products, dairy products, vegetables, fruit and related food products and animal feeds: First Action 2020.07.
McCleary, B. V. & McLoughlin, C. (2021). Journal of AOAC International, qsab019.
Background: The level of available carbohydrates in our diet is directly linked to two major diseases; obesity and Type II diabetes. Despite this, to date there is no method available to allow direct and accurate measurement of available carbohydrates in human and animal foods. Objective: The aim of this research was to develop a method that would allow simple and accurate measurement of available carbohydrates, defined as non-resistant starch, maltodextrins, maltose, isomaltose, sucrose, lactose, glucose, fructose and galactose. Method: Non-resistant (digestible) starch is hydrolysed to glucose and maltose by pancreatic α-amylase and amyloglucosidase at pH 6.0 with shaking or stirring at 37°C for 4 h. Sucrose, lactose, maltose and isomaltose are completely hydrolyzed by specific enzymes to their constituent monosaccharides, which are then measured using pure enzymes in a single reaction cuvette. Results: A method has been developed that allows the accurate measurement of available carbohydrates in all cereal, vegetable, fruit, food, and feed products, including dairy products. Conclusions: A single-laboratory validation was performed on a wide range of food and feed products. The inter-day repeatability (%RSDr) was <3.58% (w/w) across a range of samples containing 44.1 to 88.9% available carbohydrates. The LOD and LOQ obtained were 0.054% (w/w) and 0.179% (w/w), respectively. The method is all inclusive, specific, robust and simple to use. Highlights: A unique method has been developed for the direct measurement of available carbohydrates, entailing separate measurement of glucose, fructose and galactose; information of value in determining the glycemic index of foods.
Hide AbstractHighly-efficient lipid production from hydrolysate of Radix paeoniae alba residue by oleaginous yeast Cutaneotrichosporon oleaginosum.
Xu, C., Wang, Y., Zhang, C., Liu, J., Fu, H., Zhou, W. & Gong, Z. (2024). Bioresource Technology, 391, 129990.
Valorization of herbal extraction residues (HERs) into value-added products is pivotal for the sustainability of Chinese medicine industry. Here, seven different enzymatic hydrolysates of dilute acid pretreated HERs were evaluated for lipid production by Cutaneotrichosporon oleaginosum. Among them, the highest sugar yield via hydrolysis and the maximum lipid production were obtained from Radix paeoniae alba residue (RPAR). More interestingly, high proportion of sugar polymers was disintegrated into fermentable sugars during the pretreatment step, allowing a cheap non-enzymatic route for producing sugars from RPAR. A repeated dilute acid pretreatment gained a high sugar concentration of 241.6 g/L through reusing the pretreatment liquor (PL) for four times. Biomass, lipid concentration, and lipid content achieved 49.5 g/L, 35.7 g/L and 72.2 %, respectively, using fed-batch culture of PL. The biodiesel parameters indicated lipids produced from HERs were suitable for biodiesel production. This study offers a cost-effective way to upgrade the HERs waste into micro-biodiesel.
Hide AbstractXylobiose treatment triggers a defense-related response and alters cell wall composition.
Dewangan, B. P., Gupta, A., Sah, R. K., Das, S., Kumar, S., Bhattacharjee, S. & Pawar, P. A. M. (2023). Plant Molecular Biology, 113(6), 383-400.
Plant cell wall-derived oligosaccharides, i.e., damage-associated molecular patterns (DAMPs), could be generated after pathogen attack or during normal plant development, perceived by cell wall receptors, and can alter immunity and cell wall composition. Therefore, we hypothesised that xylo-oligosaccharides (XOS) could act as an elicitor and trigger immune responses. To test this, we treated Arabidopsis with xylobiose (XB) and investigated different parameters. XB-treatment significantly triggered the generation of reactive oxygen species (ROS), activated MAPK protein phosphorylation, and induced callose deposition. The combination of XB (DAMP) and flg22 a microbe-associated molecular pattern (MAMP) further enhanced ROS response and gene expression of PTI marker genes. RNA sequencing analysis revealed that more genes were differentially regulated after 30 min compared to 24 h XB-treated leaves, which correlated with ROS response. Increased xylosidase activity and soluble xylose level after 30 min and 3 h of XB-treatment were observed which might have weakened the DAMP response. However, an increase in total cell wall sugar and a decrease in uronic acid level was observed at both 30 min and 24 h. Additionally, arabinose, rhamnose, and xylose levels were increased in 30 min, and glucose was increased in 24 h compared to mock-treated leaves. The level of jasmonic acid, abscisic acid, auxin, and cytokinin were also affected after XB treatment. Overall, our data revealed that the shortest XOS can act as a DAMP, which triggers the PTI response and alters cell wall composition and hormone level.
Hide AbstractRecovery of Nanocellulose from Agri-Food Residues through Chemical and Physical Processes.
Pirozzi, A., Pappalardo, G. & Donsì, F. (2023). Chemical Engineering Transactions, 102, 175-180.
This work proposes a biorefinery approach for the exploitation of agri-food by-products, such as tomato pomace (TP), through the combination of mild chemical hydrolysis and high-pressure homogenization (HPH) in water not only to promote the recovery of cellulose but also its defibrillation to obtain nanocellulose. In particular, the cellulose pulp was isolated from TP using different combinations of chemical and physical processes, by applying HPH treatment (i) directly on the raw material, (ii) after the acid hydrolysis, and (iii) after alkaline hydrolysis. Moreover, the isolated cellulose was deconstructed to obtain cellulose nanoparticles, also through the application of the HPH treatment, enhancing the polymer properties. The structural and physical features of cellulose nanoparticles from TP were analyzed through Fourier-transform infrared spectroscopy (FT-IR) analysis, ?-potential measurement, and morphological analysis with SEM. The results clearly showed that the HPH treatment (80 MPa, 20 passes) at different stages of the process caused only a slight increase in the yield of cellulose recovery, but significantly contributed to obtaining defibrillated cellulose particles, characterized by smaller irregular domains containing elongated needle-like fibers.
Hide AbstractEngineering transcriptional regulation of pentose metabolism in Rhodosporidium toruloides for improved conversion of xylose to bioproducts.
Coradetti, S. T., Adamczyk, P. A., Liu, D., Gao, Y., Otoupal, P. B., Geiselman, G. M., Webb-Robertson, B. J. M., Burnet, M. C., Kim, Y. M., Burnum-Johnson, K. E., Magnuson, J. & Gladden, J. M. (2023). Microbial Cell Factories, 22(1), 144.
Efficient conversion of pentose sugars remains a significant barrier to the replacement of petroleum-derived chemicals with plant biomass-derived bioproducts. While the oleaginous yeast Rhodosporidium toruloides (also known as Rhodotorula toruloides) has a relatively robust native metabolism of pentose sugars compared to other wild yeasts, faster assimilation of those sugars will be required for industrial utilization of pentoses. To increase the rate of pentose assimilation in R. toruloides, we leveraged previously reported high-throughput fitness data to identify potential regulators of pentose catabolism. Two genes were selected for further investigation, a putative transcription factor (RTO4_12978, Pnt1) and a homolog of a glucose transceptor involved in carbon catabolite repression (RTO4_11990). Overexpression of Pnt1 increased the specific growth rate approximately twofold early in cultures on xylose and increased the maximum specific growth by 18% while decreasing accumulation of arabitol and xylitol in fast-growing cultures. Improved growth dynamics on xylose translated to a 120% increase in the overall rate of xylose conversion to fatty alcohols in batch culture. Proteomic analysis confirmed that Pnt1 is a major regulator of pentose catabolism in R. toruloides. Deletion of RTO4_11990 increased the growth rate on xylose, but did not relieve carbon catabolite repression in the presence of glucose. Carbon catabolite repression signaling networks remain poorly characterized in R. toruloides and likely comprise a different set of proteins than those mainly characterized in ascomycete fungi.
Hide AbstractThe effect of different wheat varieties and exogenous xylanase on bird performance and utilization of energy and nutrients.
Whiting, I. M., Pirgozliev, V. & Bedford, M. R. (2023). Poultry Science, 102817.
The aims of the present study were to first, determine the xylan fractions of 10 different wheat cultivar samples and their response to treatment by the same commercial xylanase enzyme preparation. Second, use information obtained to select 5 of the wheats for use within a feeding experiment to determine whether the rate of xylan release can be used to predict the feeding value of the wheats when diets have been supplemented with xylanase. Treatment of 10 different wheat varieties by the same enzyme resulted in varying levels of hydrolysis. Soluble xylan content ranged from 7.85 to 14.40 and 3.20 to 5.13 (mg/g) when treated with and without xylanase, respectively. Oligosaccharide content ranged from 0.34 to 1.58 and 0.05 to 0.54 (mg/g) when treated with and without xylanase, respectively. Five of the 10 wheats were then selected based on the determined xylan fractions to use within a feeding experiment. A total of 360 male Ross 308 broilers were randomly allocated to 60 raised floor pens. A soybean meal (SBM) balancer feed was formulated to contain 12.07 MJ/kg apparent metabolizable energy (AME) and 392.9 g/kg crude protein (CP). Five diets were prepared by mixing 630 g/kg of each of the 5 experimental wheats with 370 g/kg of the balancer. Each diet was split into 2, one of which was supplemented with 100 g/MT of Econase XT (223,000 BXU/g), resulting in a total of 10 diets. The birds were fed the diets from 0 to 28 d of age. Wheat cultivar had an effect (P = 0.044) on feed intake (FI), while the addition of xylanase increased (P < 0.05) weight gain (WG) and improved feed conversion ratio (FCR). Various interactions were observed (P < 0.05) between wheat cultivars and xylanase for AME and nutrient utilization. This study suggests that wheats treated with the same xylanase, differ in their susceptibility to release soluble xylan and oligosaccharides, which may partially explain the varying performance and nutrient digestibility responses noted in the literature.
Hide AbstractGreen synthesis of nickel ferrite nanoparticles for efficient enhancement of lignocellulosic hydrolysate-based biohydrogen production.
Zhang, Q., Cao, J., Zhao, P., Zhang, Y., Li, Y., Xu, S., Ye, C. & Qian, C. (2023). Biochemical Engineering Journal, 194, 108885.
To improve the lignocellulosic hydrolysate-based production of biohydrogen, bimetallic nickel ferrite nanoparticles (NPs) with small particle size, cubic shape, high stability, and biocompatibility are synthesized using an Eichhornia crassipes extract at a low annealing temperature of 400 °C and added to the lignocellulosic hydrolysate fermentation by Klebsiella sp. WL1316. The optimal addition of 30 mg/L gsNiFe2O4 NPs accounts for the highest cumulative hydrogen production of 5544.86 ± 37.03 mL/L and improvement of 112.32% ± 1.86% at 24 h, while also resulting in the highest improvement of hydrogenase and formate-hydrogen lyase activities up to 102.11% ± 13.73% and 62.99% ± 4.66% compared to the Control treatment, respectively. Moreover, the conversion efficiencies of glucose, xylose, and substrate are enhanced upon addition of gsNiFe2O4 NPs, reaching values higher than 96% in the presence of 30 mg/L gsNiFe2O4 NPs. At the same time, the hydrogen yield converted from the substrate (Y(H2/S)) and biomass converted from the substrate (Y(B/S)) are also improved. In addition, the alteration of soluble metabolic products, especially significant changes in formic acid and ethanol concentrations compared to the control, increases the flux in the formate-hydrogen lytic pathway for hydrogen evolution, thereby promoting the substrate conversion level to hydrogen gas.
Hide AbstractMonthly Variation in Mycosporine-like Amino Acids from Red Alga Dulse (Devaleraea inkyuleei, Formerly Palmaria palmata in Japan).
Yamamoto, R., Mune Mune, M. A., Miyabe, Y., Kishimura, H. & Kumagai, Y. (2023). Phycology, 3(1), 127-137.
Mycosporine-like amino acids (MAAs) are natural ultraviolet-absorbing compounds found in microalgae and macroalgae. MAA content changes seasonally and in response to environmental factors. We previously investigated MAAs from the red alga dulse (Devaleraea inkyuleei, formerly Palmaria palmata in Japan) in Usujiri, Hokkaido, Japan, from 2019 to 2020. At that time, some factors affecting MAA content were still unclear. In this study, we investigated MAA variation during the period from January to June 2021, and evaluated new methods of MAA extraction from dulse. We recorded a maximum MAA extraction yield (7.03 µmol/g dry weight) on 25 March 2021. Over the course of our three years of investigations from 2019 to 2021, we found that dulse was most suitable for MAA preparation from the middle of February to late April. In the later work reported in this paper, we improved our extraction method by using a lower-risk organic solvent (ethanol) rather than methanol. In addition, we evaluated MAA extraction using different levels of ethanol concentration (25, 50, and 99%) and different extraction times (2, 6, and 24 h). We found that extraction with 25% ethanol for 24 h increased MAA content by a factor of 3.2, compared with our previous extraction method. In summary, we determined the most suitable sampling period for Usujiri dulse, to extract the highest content of MAAs. We also improved the effectiveness of the extraction process.
Hide AbstractHighly efficient fed-batch modes for enzymatic hydrolysis and microbial lipogenesis from alkaline organosolv pretreated corn stover for biodiesel production.
Wang, X., Wang, Y., He, Q., Liu, Y., Zhao, M., Liu, Y., Zhou, W. & Gong, Z. (2022). Renewable Energy, 197, 1133-1143.
High-density culture for microbial lipid preparation from low-cost lignocellulosic feedstocks is crucial for commercial-scale biodiesel production. Herein, fed-batch saccharification of alkaline organosolv pretreatment (AOP) of corn stover at an extremely high solids content of 47% (w/v) released 299.5 g/L of lignocellulosic sugars including 18.3% of soluble oligosaccharides. Three types of liquid hydrolysates for seed culture, fermentation, and feeding during fed-batch culture were obtained from the hydrolysate slurry using a two-step washing strategy with 99.3% of sugars recovery. Cutaneotrichosporon oleaginosum showed excellent capacity for assimilating both monosaccharides and oligosaccharides for lipid production using the fed-batch culture mode. Lipid concentration, content, and yield gained 42.3 g/L, 64.6%, and 20.4 g/100 g, respectively. Turbid hydrolysate collected with high recovery of high-concentration sugars and simplified process could be directly served as feeding medium. In general, the overall hydrolysis yield and lipid yield using fed-batch mode accounted for 93.2% and 97.6% of those using batch mode, respectively, resulting in a lipid output of 102.8 g/kg raw corn stover. The fatty acid composition and the prediction of biodiesel properties of lipid samples indicated the suitability for high-quality fuel production. This study provided valuable information for designing highly efficient lignocelluloses-to-biodiesel routes.
Hide AbstractComplete genome sequencing and identification of fiber-degrading potential in Bacillus amyloliquefaciencs strain TL106 from the Tibetan pig.
Shang, Z. D., Liu, S., Duan, Y., Bao, C., Wang, J., Dong, B. & Cao, Y. H. (2022). BMC Microbiol, 22(1),186.
Background: Cellulolytic microorganisms are considered a key player in the degradation of feed fiber. These microorganisms can be isolated from various resources, such as animal gut, plant surfaces, soil and oceans. A new strain of Bacillus amyloliquefaciens, TL106, was isolated from faeces of a healthy Tibetan pigs. This strain can produce cellulase and shows strong antimicrobial activity in mice. Thus, in this study, to better understand the strain of B. amyloliquefaciens TL106 on degradation of cellulose, the genome of the strain TL106 was completely sequenced and analyzed. In addition, we also explored the cellulose degradation ability of strain TL106 in vitro. Results: TL106 was completely sequenced with the third generation high-throughput DNA sequencing. In vitro analysis with enzymatic hydrolysis identified the activity of cellulose degradation. TL106 consisted of one circular chromosome with 3,980,960 bp and one plasmid with 16,916 bp, the genome total length was 3.99 Mb and total of 4,130 genes were predicted. Several genes of cellulases and hemicellulase were blasted in Genbank, including β-glucosidase, endoglucanase, ß-glucanase and xylanase genes. Additionally, the activities of amylase (20.25 U/mL), cellulase (20.86 U/mL), xylanase (39.71 U/mL) and β-glucanase (36.13 U/mL) in the fermentation supernatant of strain TL106 were higher. In the study of degradation characteristics, we found that strain TL106 had a better degradation effect on crude fiber, neutral detergent fiber, acid detergent fiber, starch, arabinoxylan and β-glucan of wheat and highland barley . Conclusions: The genome of B. amyloliquefaciens TL106 contained several genes of cellulases and hemicellulases, can produce carbohydrate-active enzymes, amylase, cellulase, xylanase and β-glucanase. The supernatant of fermented had activities of strain TL106. It could degrade the fiber fraction and non-starch polysaccharides (arabinoxylans and β-glucan) of wheat and highland barley. The present study demonstrated that the degradation activity of TL106 to crude fiber which can potentially be applied as a feed additive to potentiate the digestion of plant feed by monogastric animals.
Hide AbstractChemical composition and bioactivity of oilseed cake extracts obtained by subcritical and modified subcritical water.
Švarc-Gajić, J., Rodrigues, F., Moreira, M. M., Delerue-Matos, C., Morais, S., Dorosh, O., Silva, A. M., Bassani, A., Dzedik, V. & Spigno, G. (2022). Bioresources and Bioprocessing, 9(1), 1-14.
Recovery of bioactive compounds from biowaste is gaining more and more interest in circular economy models. The oilseed cakes are usually insufficiently exploited by most technologies since they represent valuable matrices abundant in proteins, minerals, and phytochemicals, but their use is mostly limited to feed ingredients, fertilizers or biofuel production. This study was thus focused on the exploration of new valorization pathways of oilseed cakes by subcritical water, representing a safe and economic alternative in the creation of value chains. Pumpkin, hemp, and flax seed cakes were treated with subcritical water in nitrogen and carbon-dioxide atmospheres, as well as in nitrogen atmosphere with the addition of acid catalyst. The degradation of carbohydrate fraction was studied by quantifying sugars and sugar degradation products in the obtained extracts. The extracts obtained under different conditions were further compared chemically with respect to total phenols and flavonoids, as well as to the content of individual phenolic compounds. Furthermore, the effects of subcritical water treatment conditions on antioxidant, antiradical and cytotoxic properties of thus obtained extracts were defined and discussed.
Hide AbstractArabidopsis GELP7 functions as a plasma membrane-localized acetyl xylan esterase, and its overexpression improves saccharification efficiency.
Rastogi, L., Chaudhari, A. A., Sharma, R. & Pawar, P. A. M. (2022). Plant Molecular Biology, 109, 781-797.
Acetyl substitution on the xylan chain is critical for stable interaction with cellulose and other cell wall polymers in the secondary cell wall. Xylan acetylation pattern is governed by Golgi and extracellular localized acetyl xylan esterase (AXE). We investigated the role of Arabidopsis clade Id from the GDSL esterase/lipase or GELP family in polysaccharide deacetylation. The investigation of the AtGELP7 T-DNA mutant line showed a decrease in stem esterase activity and an increase in stem acetyl content. We further generated overexpressor AtGELP7 transgenic lines, and these lines showed an increase in AXE activity and a decrease in xylan acetylation compared to wild-type plants. Therefore, we have named this enzyme as AtAXE1. The subcellular localization and immunoblot studies showed that the AtAXE1 enzyme is secreted out, associated with the plasma membrane and involved in xylan de-esterification post-synthesis. The cellulose digestibility was improved in AtAXE1 overexpressor lines without pre-treatment, after alkali and xylanases pre-treatment. Furthermore, we have also established that the AtGELP7 gene is upregulated in the overexpressor line of AtMYB46, a secondary cell wall specific transcription factor. This transcriptional regulation can drive AtGELP7 or AtAXE1 to perform de-esterification of xylan in a tissue-specific manner. Overall, these data suggest that AtGELP7 overexpression in Arabidopsis reduces xylan acetylation and improves digestibility properties of polysaccharides of stem lignocellulosic biomass.
Hide AbstractHigh-pressure autohydrolysis process of wheat straw for cellulose recovery and subsequent use in PBAT composites preparation.
Fiorentini, C., Bassani, A., Garrido, G. D., Merino, D., Perotto, G., Athanassiou, A., Prantie, J., Halonen, N. & Spigno, G. (2022). Biocatalysis and Agricultural Biotechnology, 39, 102282.
The effect of autohydrolysis (AH) temperature (165°C, 195°C, 225°C) on the structure, purity, and recovery yield of the cellulose residue isolated after additional alkaline and bleaching steps from wheat straw, was investigated. The processes were quantified for mass yields in the different steps and for antioxidants and sugars release during AH. AH at 195°C allowed for the highest cellulose residue yield (83.5%) with purity (~70%) and structure similar to the other residues. FTIR and XRD analyses showed straw cellulose (SC) with a type II polymorphism and crystallinity index increasing with AH temperature. SC obtained at the end of the entire fractionation process (SC-195°C) starting from AH residue-195°C was tested as a reinforcing agent in different percentage (0, 2 and 5% by weight) in poly(butylene adipate-co-terephthalate) (PBAT) films. The Young's modulus of the films increased by ~17% with 5 wt% cellulose, while tensile strength and elongation at break decreased.
Hide AbstractAbility of yeast metabolic activity to reduce sugars and stabilize betalains in red beet juice.
Dygas, D., Nowak, S., Olszewska, J., Szymańska, M., Mroczyńska-Florczak, M., Berłowska, J., Dziugan, P. & Kręgiel, D. (2021). Fermentation, 7(3), 105.
To lower the risk of obesity, diabetes, and other related diseases, the WHO recommends that consumers reduce their consumption of sugars. Here, we propose a microbiological method to reduce the sugar content in red beet juice, while incurring only slight losses in the betalain content and maintaining the correct proportion of the other beet juice components. Several yeast strains with different metabolic activities were investigated for their ability to reduce the sugar content in red beet juice, which resulted in a decrease in the extract level corresponding to sugar content from 49.7% to 58.2%. This strategy was found to have the additional advantage of increasing the chemical and microbial stability of the red beet juice. Only slight losses of betalain pigments were noted, to final concentrations of 5.11% w/v and 2.56% w/v for the red and yellow fractions, respectively.
Hide AbstractCitrulline supplementation attenuates the development of non-alcoholic steatohepatitis in female mice through mechanisms involving intestinal arginase.
Rajcic, D., Baumann, A., Hernández-Arriaga, A., Brandt, A., Nier, A., Jin, C. J., Sanchez, V., Jumg, F., Camarinha-Silva, A. & Bergheim, I. (2021). Redox Biology, 41, 101879.
Non-alcoholic fatty liver disease (NAFLD) is by now the most prevalent liver disease worldwide. The non-proteogenic amino acid l-citrulline (L-Cit) has been shown to protect mice from the development of NAFLD. Here, we aimed to further assess if L-Cit also attenuates the progression of a pre-existing diet-induced NAFLD and to determine molecular mechanisms involved. Female C57BL/6J mice were either fed a liquid fat-, fructose- and cholesterol-rich diet (FFC) or control diet (C) for 8 weeks to induce early stages of NASH followed by 5 more weeks with either FFC-feeding +/- 2.5 g L-Cit/kg bw or C-feeding. In addition, female C57BL/6J mice were either pair-fed a FFC +/- 2.5 g L-Cit/kg bw +/- 0.01 g/kg bw i.p. N(ω)-hydroxy-nor-l-arginine (NOHA) or C diet for 8 weeks. The protective effects of supplementing L-Cit on the progression of a pre-existing NAFLD were associated with an attenuation of 1) the increased translocation of bacterial endotoxin and 2) the loss of tight junction proteins as well as 3) arginase activity in small intestinal tissue, while no marked changes in intestinal microbiota composition were prevalent in small intestine. Treatment of mice with the arginase inhibitor NOHA abolished the protective effects of L-Cit on diet-induced NAFLD. Our results suggest that the protective effects of L-Cit on the development and progression of NAFLD are related to alterations of intestinal arginase activity and intestinal permeability.
Hide Abstract